OMERO
- Servers
- Uploading data
- Fix Cellomics Plate indexing
- Modifying contrast settings
- Creating figures with OMERO Figure
- Downloading a complete plate
- Tagging imaging errors
- Listing plate images for CellProfiler import
- Importing data to CellProfiler
Servers
We have two servers running on CSC ePouta. Currently both are considered to be only for testing, not for permanent storage of data or annotations.
- csc-lmu-omero-2.biocenter.helsinki.fi (5.3.x, works with CellProfiler 2.2.x and 3.0.x)
In the long run, the idea is to make csc-lmu-omero-1 the production server, and use the other one for testing new versions.
You can log in with your AD account, or using a shared test account (username: tester, password: testing123).
Uploading data
To upload data you need OMERO-insight client software (see instructions). It is available in the university Software Portal and already installed on the following LMU instruments:
- CellInsight
Fix Cellomics Plate indexing
There is a bug in OMERO that causes the plate columns shift to the right by one (see http://trac.openmicroscopy.org/ome/ticket/13230). Until the bug is fixed, there is a temporary solution to fix the view.
- Select the plate.
- Click Scripts → Fix Cellomics Plate Index...
- Verify the plate id
- Click OK.
This fix is incomplete, because it doesn't recover data in the last column that was pushed out of the view. These images can be found in "Orphaned images".
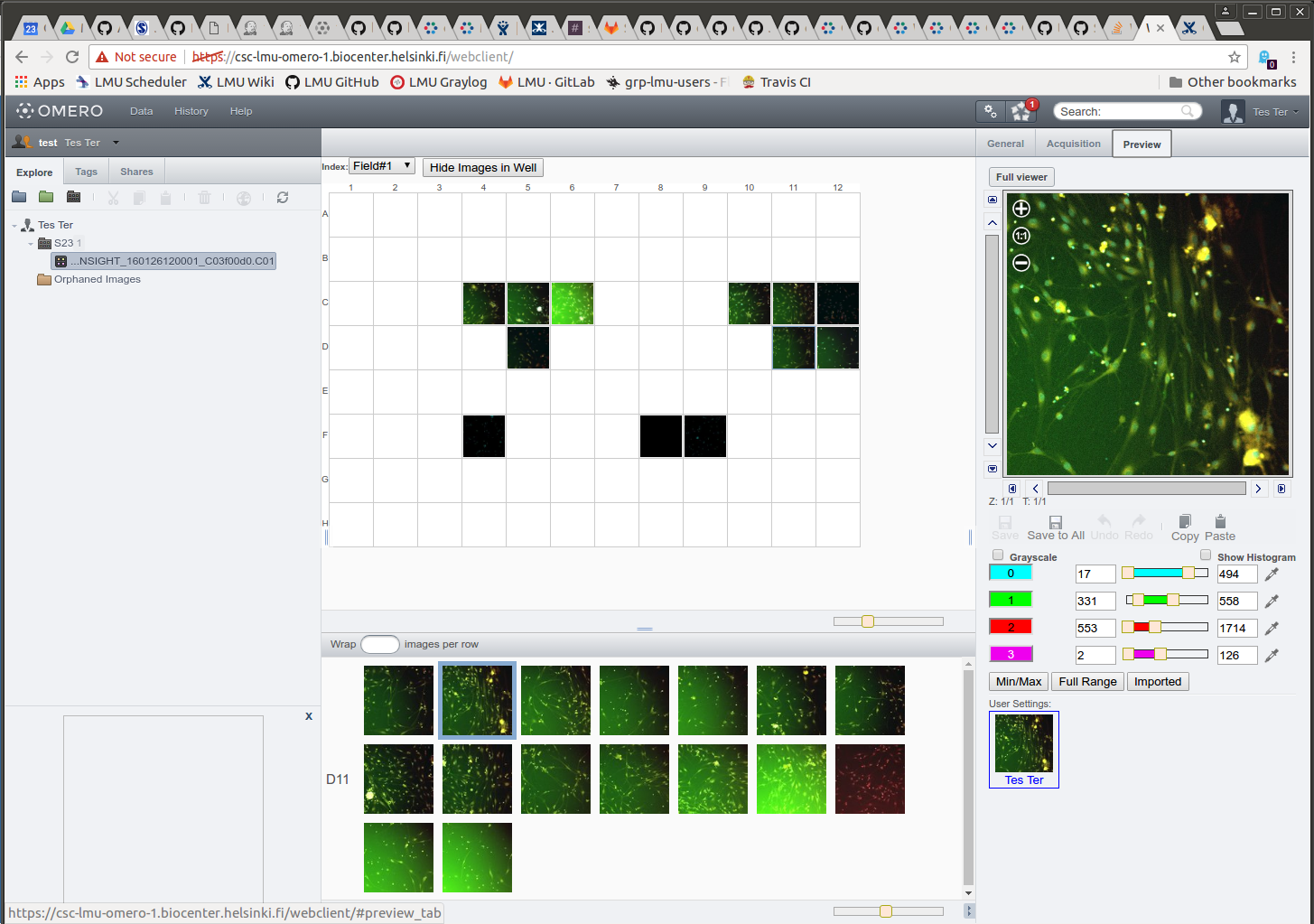
Modifying contrast settings
- Select well.
- Select field.
- Open Preview tab.
- Modify channel colors and contrast settings.
- Click "Save to All" to apply same settings to the plate.
Creating figures with OMERO Figure
If you need better quality figures, you can try the OMERO Figure application.
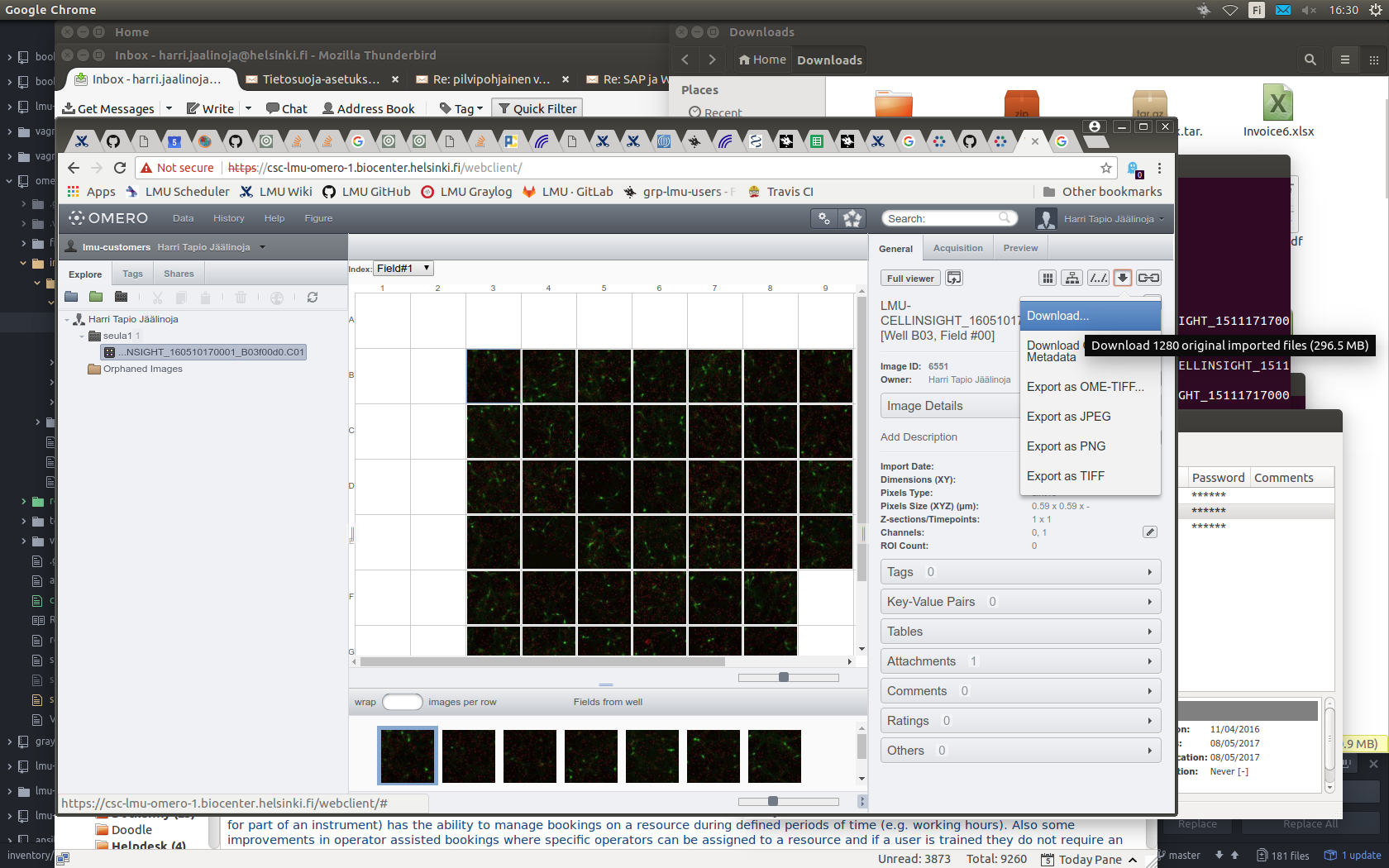
Downloading a complete plate
- Select plate.
- Select well.
- Select image.
- Click the "Download" arrow.
- Select "Download".
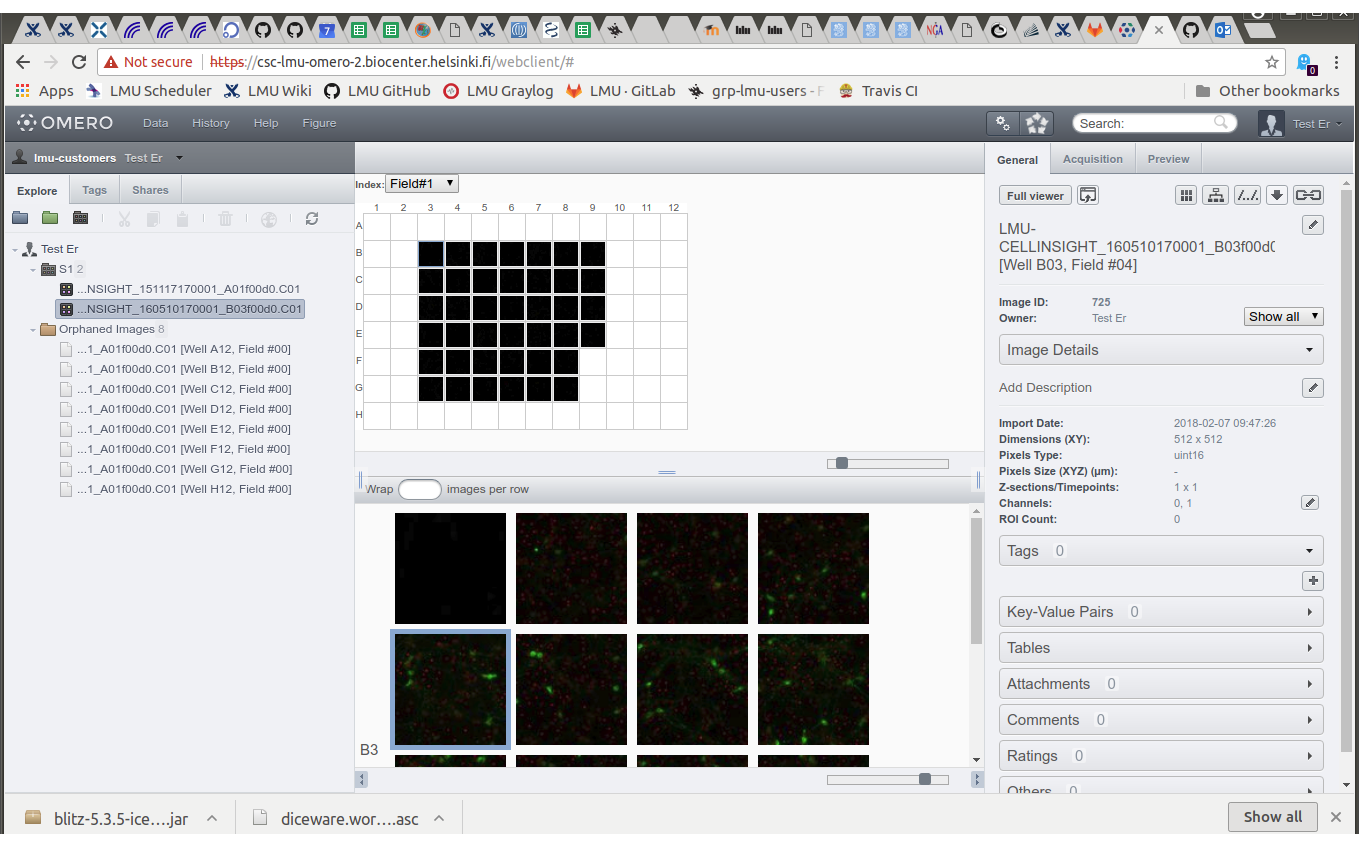
Tagging imaging errors
Especially the CellInsight instrument at LMU is plagued with imaging errors. We can use tags to mark bad images, and exclude tagged images from image lists.
- Select plate.
- Select well.
- Select image.
- On the right pane, expand Tags and click "+".
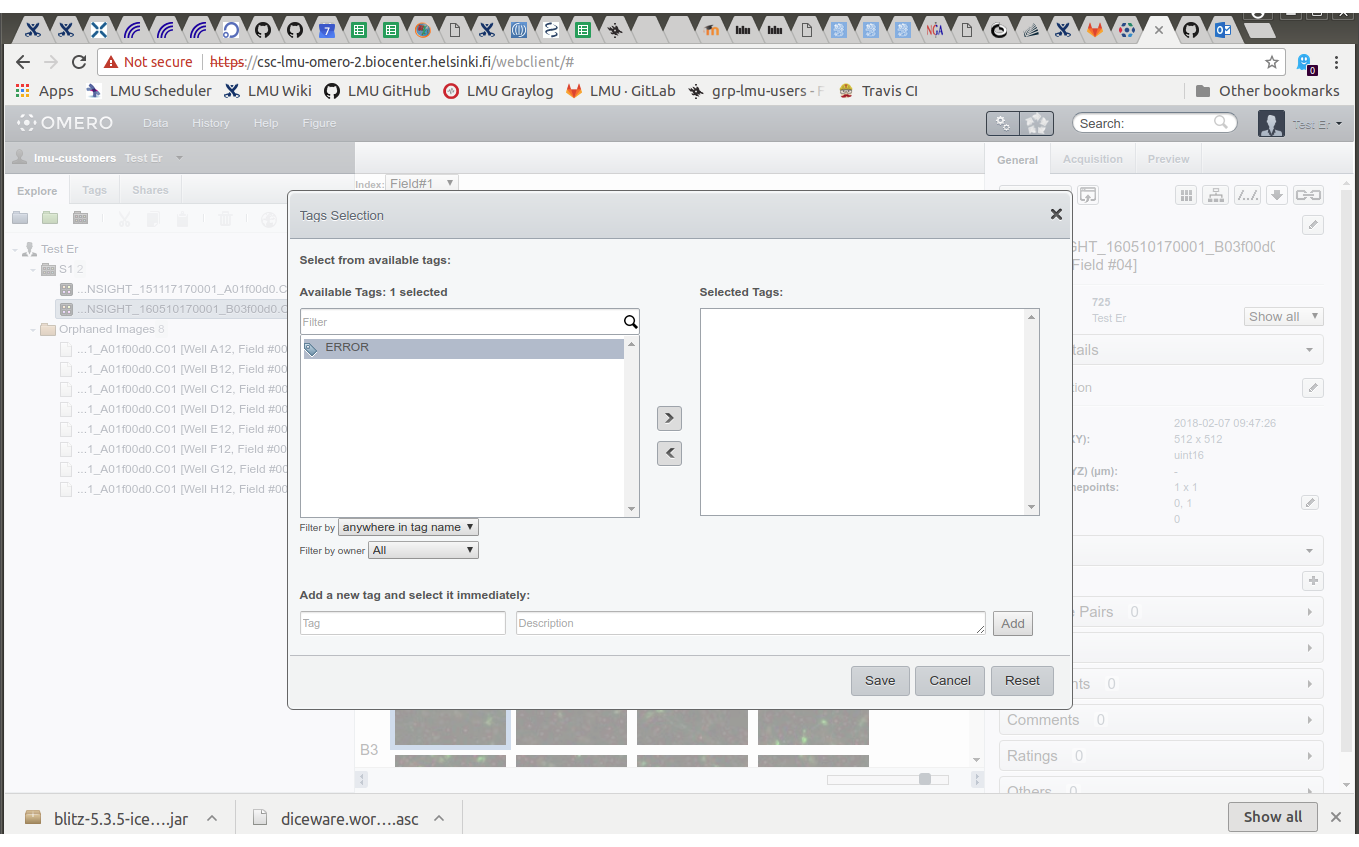
- Select the tag "ERROR", click ">", click "Save"
Listing plate images for CellProfiler import
CellProfiler can get the images directly from an OMERO server. For this purpose, we need to list the images we want to process.
- Select plate, note plate index.
- Click script icon, select "list plate images...".
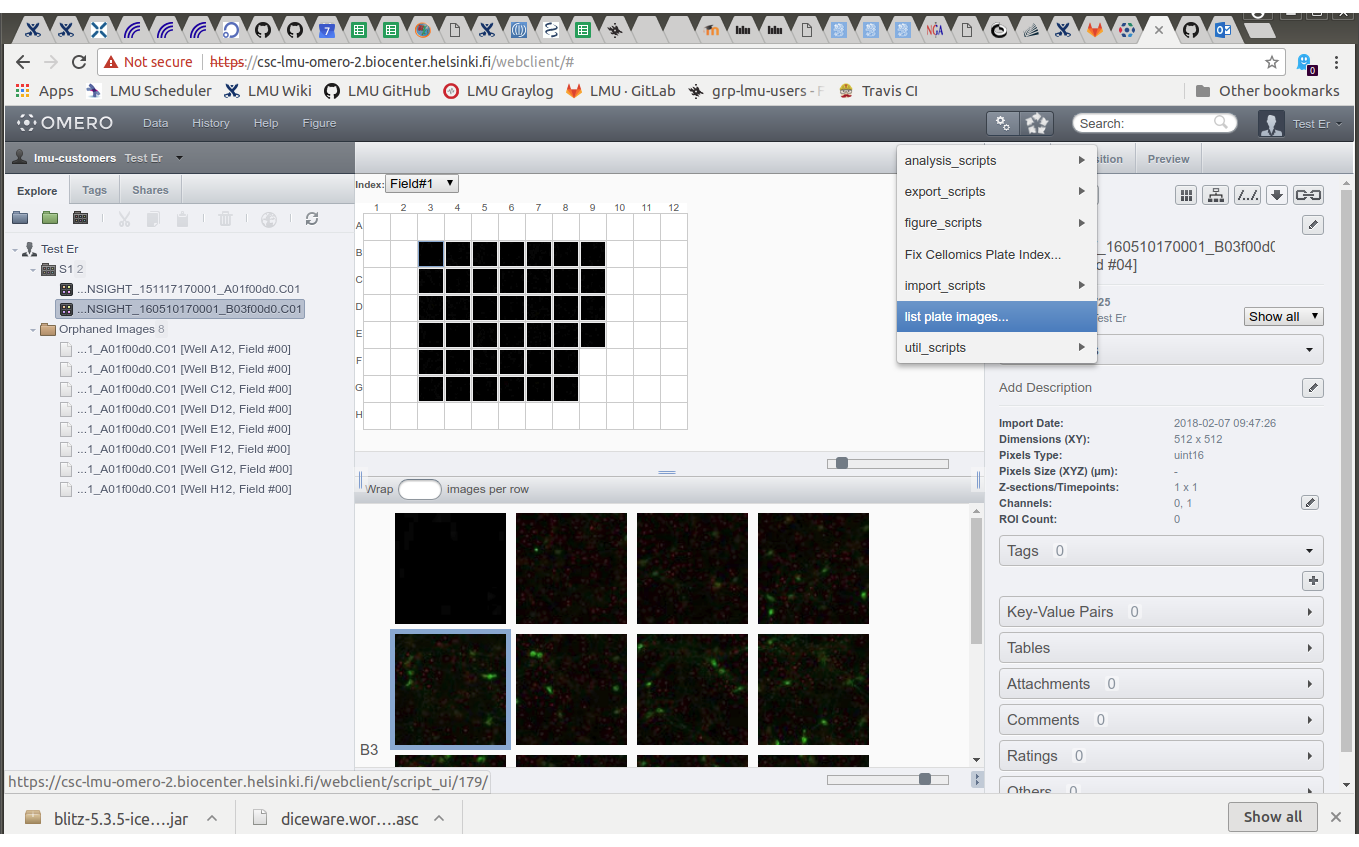
- Type the plate index and click "Run Script".
- On the Activities list, click info icon "i" on the item created by the script.
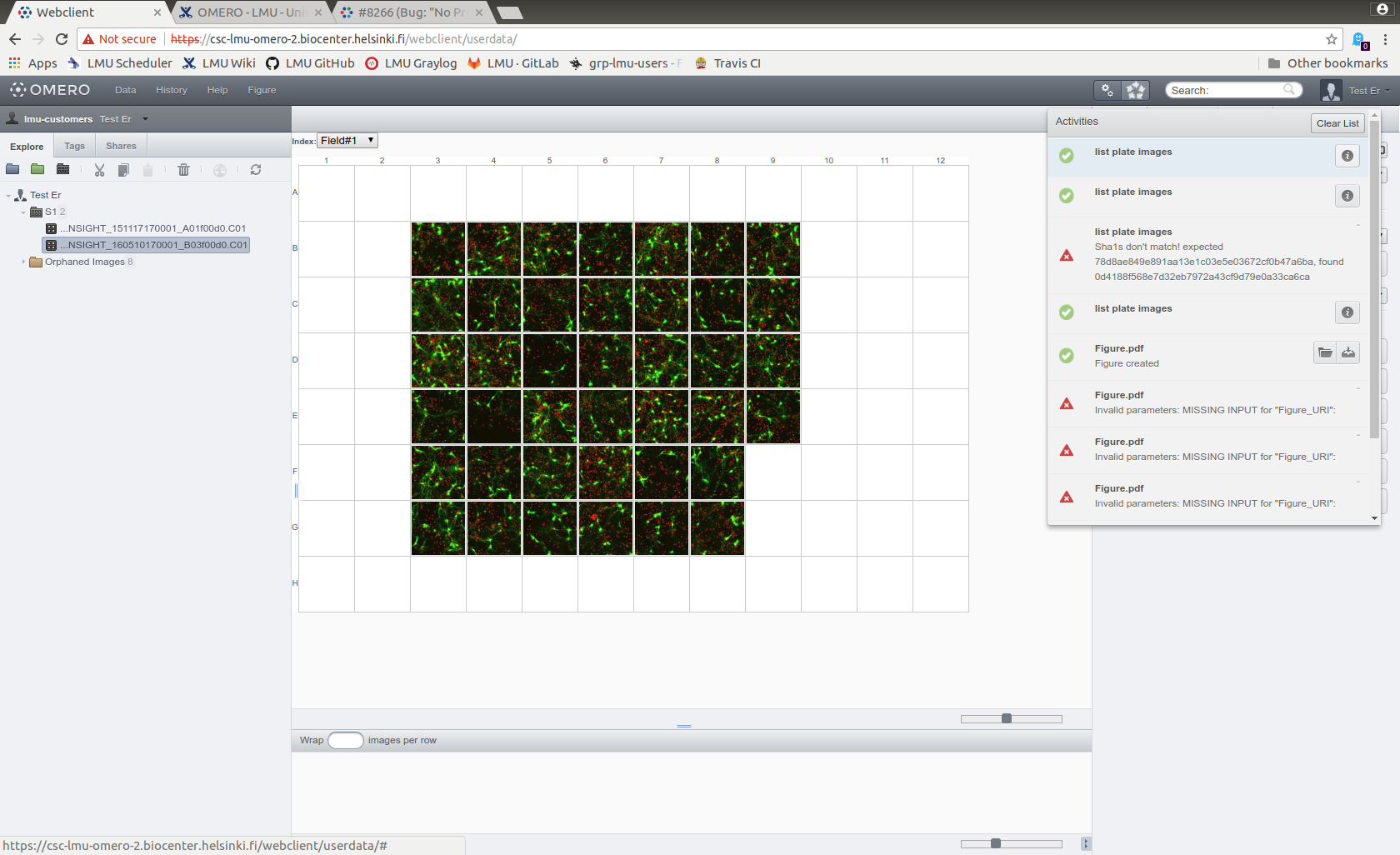
- A new window with a file list will open, cut and paste the list to a text file.
Images tagged with "ERROR" are excluded from the list.
Importing data to CellProfiler
Server csc-lmu-omero-2.biocenter.helsinki.fi is running OMERO version 5.3.x, and that has been tested to work together with CellProfiler versions 2.2.0, 2.3.2 and 3.0.* (for details see https://github.com/CellProfiler/prokaryote/issues/33).
To import the data in CellProfiler, see https://github.com/CellProfiler/CellProfiler/wiki/OMERO:-Accessing-images-from-CellProfiler. In short, you can use the LoadData module.
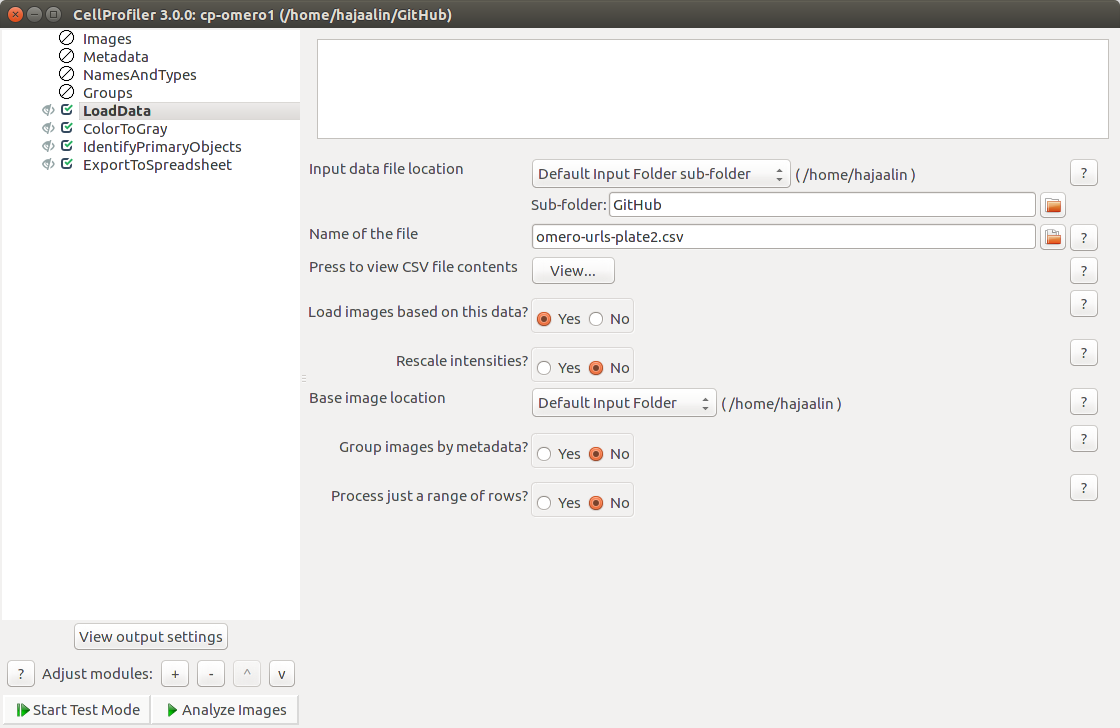
CellProfiler will ask you for your OMERO credentials, but seems to forget them after some time. If the protocol first works, but then stops working, try restarting CellProfiler.