NeighborNet
The NeighborNet algorithm is an algorithm based on neighbour joining to construct phylogenetic networks. Like neighbor joining, this method takes a distance matrix as input and starts with a completely unresolved tree (i.e. a tree connecting all nodes individually to a single root). It determines which two edges (or taxa/branches) may be connected to a new intermediate node because they are the least distant ones compared to any of the other branch combinations in the network. This process is repeated until there are no 'unprocessed' edges left. The difference to neighbour joining is that NeighborNet allows for collections of clusters that overlap and do not form a hierarchy. The SplitsTree software package implements several phylogenetic algorithms amongst which NeigborNet.
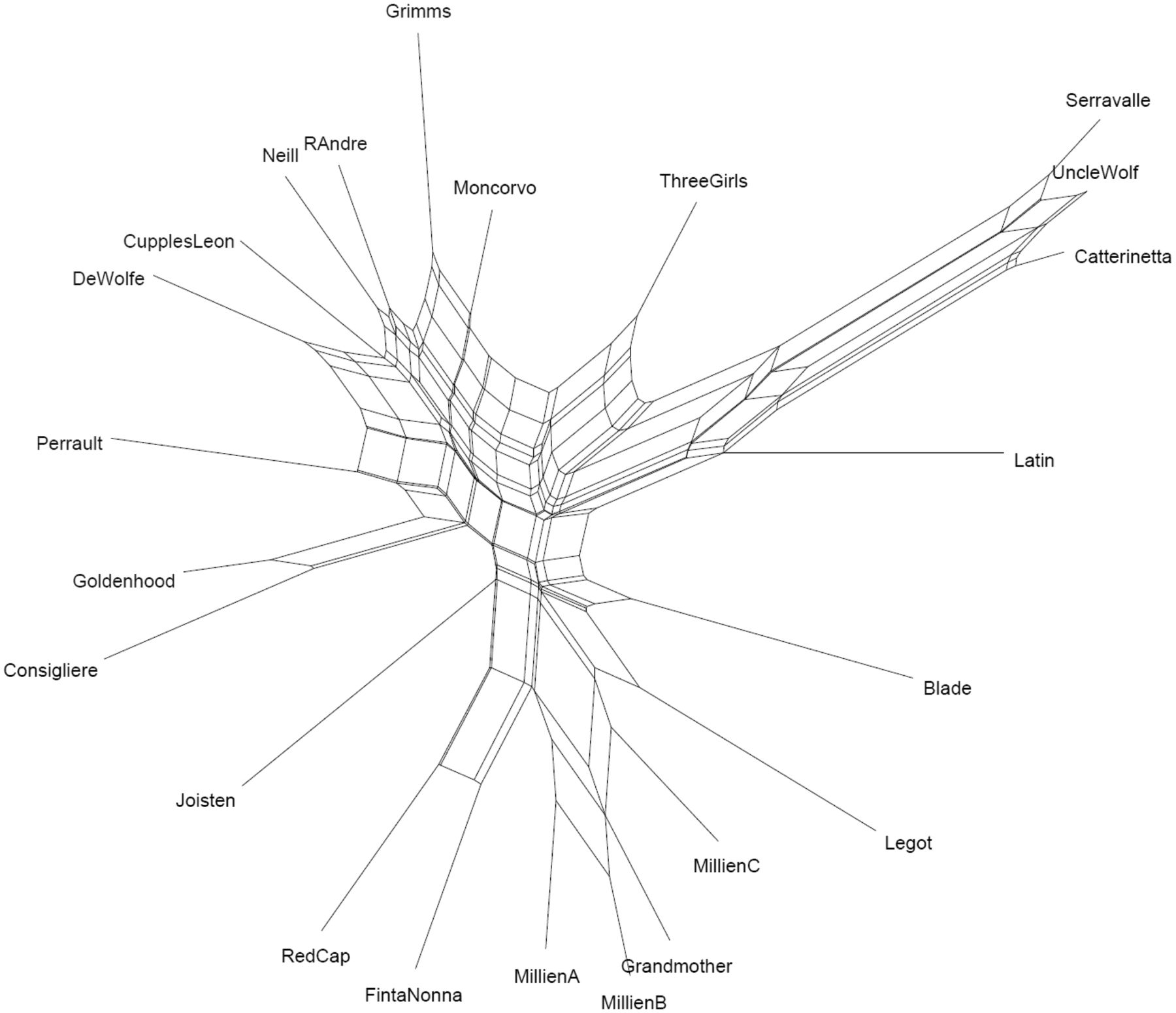
Fig. 1: Neighbornet representation of different versions of Red Ridinghood (Tehrani et al. 2015)
References
– Bryant, David, and Vincent Moulton. 2004. “Neighbor-Net: An Agglomerative Method for the Construction of Phylogenetic Networks.” Molecular Biology and Evolution 21 (2): 255–265.
– Huson, Daniel H., and David Bryant. 2006. “Application of Phylogenetic Networks in Evolutionary Studies.” Molecular Biology and Evolution 23 (2): 254–267.
– Tehrani, Jamie, Quan Nguyen, and Teemu Roos. 2015. “Oral fairy tale or literary fake? Investigating the origins of Little Red Riding Hood using phylogenetic network analysis.” Digital Scholarship in the Humanities. Advance Access 23 June 2015: http://dsh.oxfordjournals.org/content/early/2015/06/21/llc.fqv016.abstract.