Phylogenetic tree
A phylogenetic tree on a set X is a tree whose set of leaves is equal to X and which is meant to represent the phylogenetic relatedness between these objects in X. In biology these objects tend to be organisms or groups of organisms (such as species), in textual criticism they are witnesses of a text. Usually such trees are rooted and the root represents the last common ancestor (in textual criticism called the archetype). For mathematical convenience it is often assumed that there are no interior nodes with degree two since otherwise an arbitrary number of equivalent trees could be obtained by adding degree two nodes along any edge in the tree (see, e.g. Aluru 2005). Note that this is a technical requirement and it does not suggest that, in terms of textual criticism, there could not exist preserved witnesses that have exactly one descendant.
Illustration
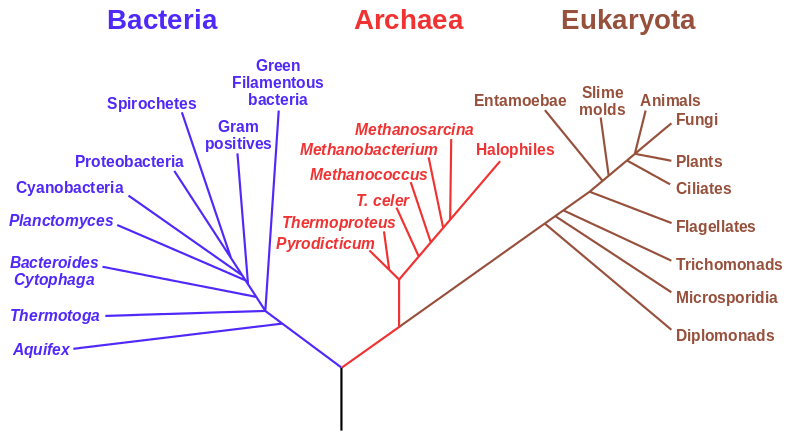
Fig. 1. An example of a phylogenetic tree of the major groups of life forms, based on RNA data and proposed by Carl Woese. (Source: https://en.wikipedia.org/wiki/File:Phylogenetic_tree.svg)
Reference
– Srinivas Aluru, ed. 2005. Handbook of Computational Molecular Biology. Boca Raton, Florida: Chapman & Hall/CRC.
In other languages
DE: phylogenetischer Baum
FR: arbre phylogénétique
IT: albero filogenetico